Why Study the Neural Crest?
Our goal is to understand how transcription factors regulate cell fate decisions in neural crest cells and to develop and study patient-specific animal models for craniofacial syndromes
One of the main goals in our lab is to understand how duplication and divergence of transcription factors leads to the formation of new cell types. The neural crest is an ideal embryonic cell population to study this as the evolutionary emergence of this cell type correlated with the duplication of many groups of transcription factors.
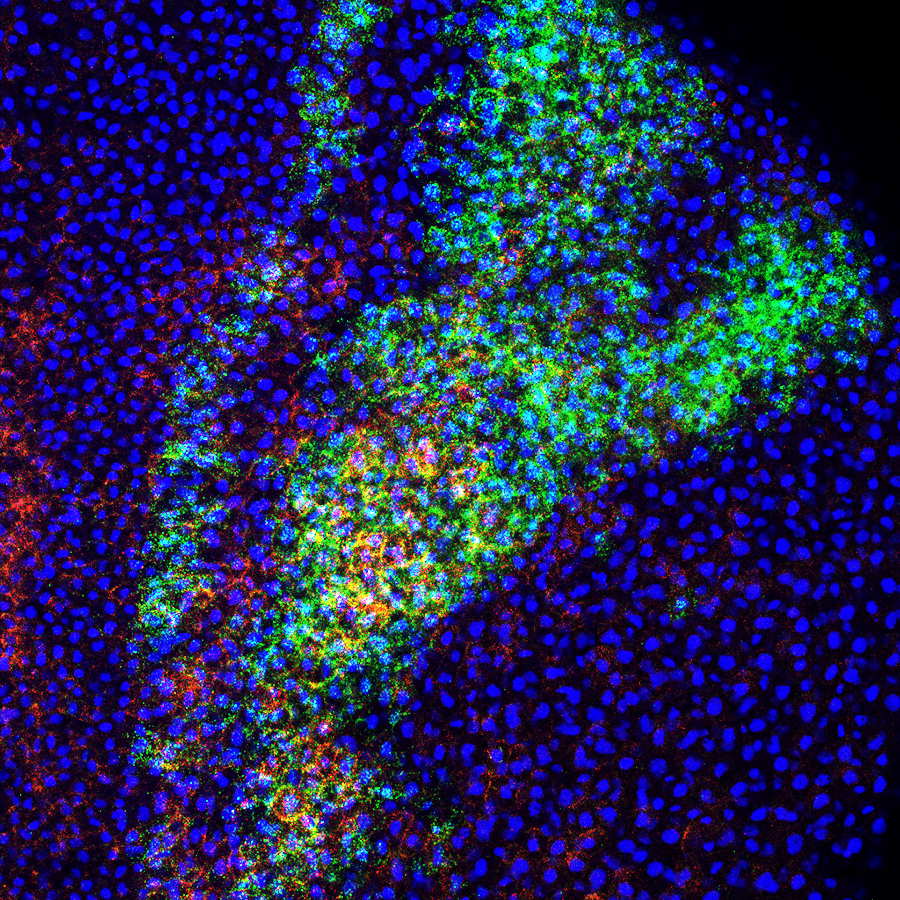
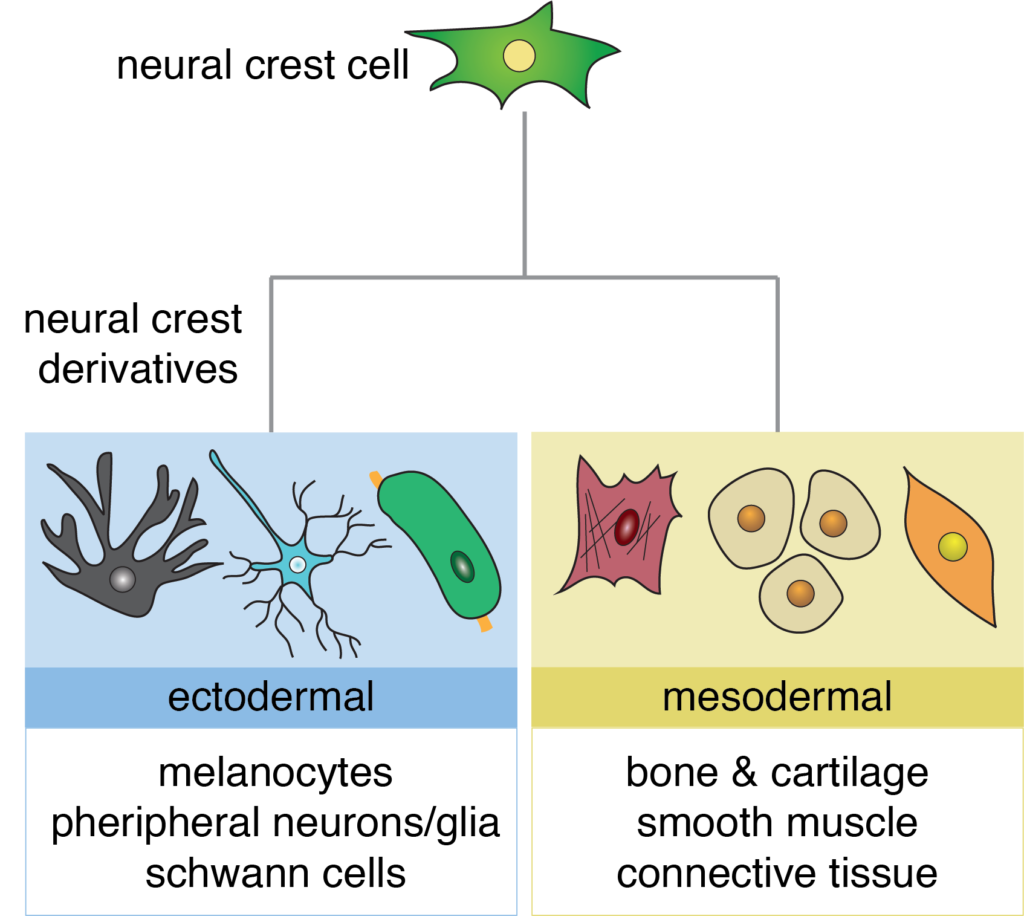
Neural crest cells are one of the most fascinating cell types in the embryo. They are an ectodermal cell population that can give rise cell types associated with both the ectoderm (pigment cells, peripheral neurons/glia) and the mesoderm (bone, cartilage, smooth muscle). Due to the vast number of derivatives with diverse functions, neural crest cells are an excellent cell type to study how transcription factors direct cell fate decisions.
Neural Crest Differentiation
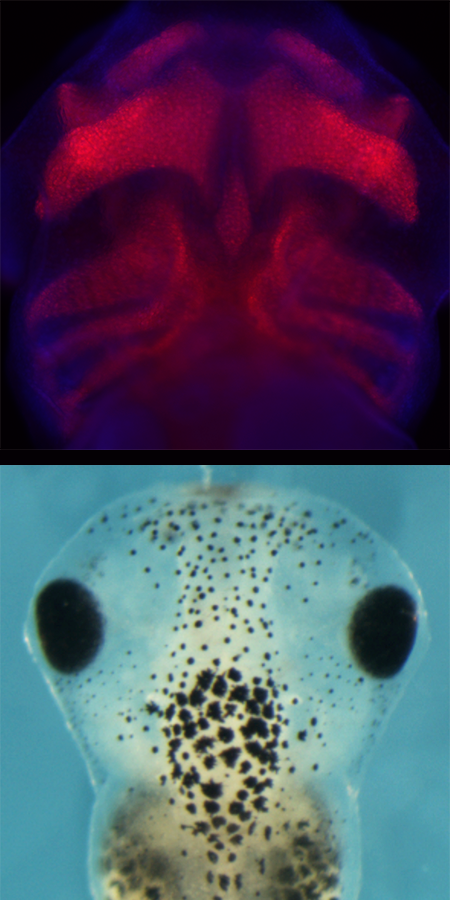
SoxE transcription factors (Sox 9 and Sox10) are essential for neural crest cell formation and differentiation. While Sox9 and Sox10 function redundantly during neural crest cell formation, they take on distinct roles as neural crest differentiate into different lineages. Sox9 directs neural crest cells to become cartilage cells while Sox10 coordinates the formation of melanocytes (pigment cells) and peripheral glia. The Schock lab is interested in understanding how transcription factors from the same subfamily, with highly similar DNA binding domains, direct differentiation of the same cell type into different lineages.
We are using Xenopus to:
- Determine how sequence differences in Sox9 and Sox10 contribute to functional differences between these factors during neural crest differentiation through Sox9-Sox10 chimera constructs and functional and genomic occupancy studies
- Assess when Sox9 and Sox10 switch from interacting with shared target genes to different target genes involved in lineage-specialization in neural crest cells through ChIP-seq
- Investigate if levels of Sox9 to Sox10 in early neural crest cells bias individual/sub-populations of cells towards specific-lineages through use of tagged enhancer-constructs and chromatin accessibility assays
Modeling Human Disease
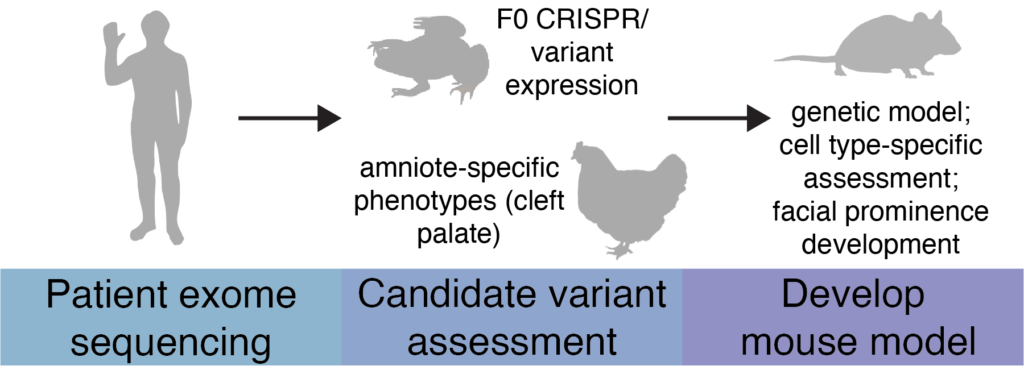
Animal models provide significant insights into human disease, including understanding syndromes with craniofacial abnormalities. A major limitation of traditional applications of model system studies, however, is that they fail to capture the complexity of human disease, as syndromic phenotypes vary from patient-to-patient. Current approaches to understanding disease include gain- and loss-of-function studies for causative genes, but often such studies are too broad, failing to make strong efforts into understanding specific genotype-phenotype correlations.
In the Schock lab, we will utilize multiple model organisms, Xenopus, chick, and mice, to investigate patient specific-variants associated with congenital craniofacial syndromes. Xenopus are an excellent and inexpensive model system for rapidly screening genes of interest and for assaying how patient-specific variants impact formation of the face. We will use Xenopus to find genes that recapitulate patient phenotypes and aggressively investigate gene function during development. If a patient phenotype is unable to be assessed in Xenopus (cleft palate) we will use chick to screen the gene instead. Once we have validated a patient variant as being causative for a syndrome, we will develop a parallel mouse model where we will study how amniote-specific aspects of craniofacial development, including facial patterning, signaling center establishment, and development of amniote-specific craniofacial structures (ex. palate) are impacted by a patient variant.
We will begin by modeling SOX9 and SOX10 variants present in Campomelic dysplasia and Waardenburg syndrome patients, respectively. We have also established a collaboration with Genomics Board at Yale where we will receive craniofacial patient portfolios and results of exome sequencing. These will be the basis of future projects in the lab.